PDF) Selection footprints in barley breeding lines detected by combining genotyping-by-sequencing with reference genome information | Elsayed Mansour - Academia.edu

Cross transferability of barley nuclear SSRs to pearl millet genome provides new molecular tools for genetic analyses and marker assisted selection
Identification and mapping of resistance genes against soil-borne viruses in barley (Hordeum vulgare L.) and wheat (Triticum aes
UNDERSTANDING EPISTASIS IN LINKAGE ANALYSIS: THE Kap AND LOCI IN THE OREGON WOLFE BARLEY POPULATION Serena B. McCoy Bachelor of

Understanding epistasis in linkage analysis : the Kap and lks2 loci in the Oregon Wolfe Barley poputlation | Semantic Scholar

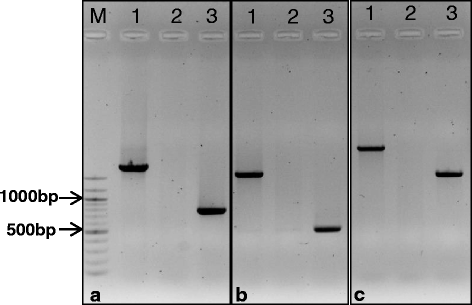
Banding pattern of the nSSR loci HVM54 and HVM03 for two barley (B1 and... | Download Scientific Diagram
MICROSATELLITE ANALYSIS OF GENETIC DIVERSITY OF WILD BARLEY (HORDEUM VULGARE SUBSP. SPONTANEUM) USING DIFFERENT SAMPLING METHODS

Genomics-based high-resolution mapping of the BaMMV/BaYMV resistance gene rym11 in barley (Hordeum vulgare L.) | SpringerLink

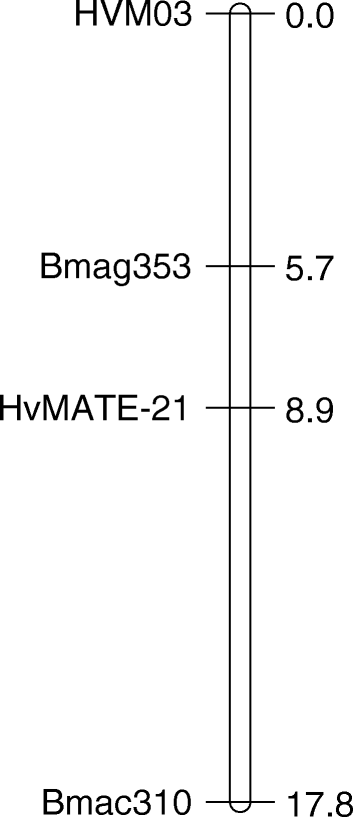
Linkage map of the gene-specific marker Cit7 on chromosome 4H in the... | Download Scientific Diagram

PDF) High genetic diversity revealed in barley ( Hordeum vulgare ) collected from small-scale farmer ' s fields in | Jihad Orabi - Academia.edu
Molecular Diversity in Bulgarian and Cypriot Barley Germplasm Collections – A Reference Point for Better Understanding, Exploi

PDF) Development of gene-specific markers for acid soil/aluminium tolerance in barley (Hordeum vulgare L.) | Meixue Zhou - Academia.edu
Analysis of the efficiency in the Spanish National Barley Breeding Program. Past results and prospects for future improvements u

Banding pattern of the nSSR loci HVM54 and HVM03 for two barley (B1 and... | Download Scientific Diagram

Understanding epistasis in linkage analysis : the Kap and lks2 loci in the Oregon Wolfe Barley poputlation | Semantic Scholar
Identification and mapping of resistance genes against soil-borne viruses in barley (Hordeum vulgare L.) and wheat (Triticum aes

Characterization of the ten transferable microsatellite markers across... | Download Scientific Diagram